class: center, middle, inverse, title-slide .title[ # Lecture 6: Sequence evolution ] .subtitle[ ## BE_22 Bioinformatics SS 21 ] .author[ ### January Weiner ] .date[ ### 2024-05-13 ] --- .pull-left[ </br> </br> <div style="font-size:150%"> <i>Nothing in biology makes sense except in the light of evolution</i> <div style="text-align:right;">– Theodosius Dobzhansky</div> </div> ] .pull-right[  ] ??? "light of evolution" (sub specie evolutionis) was used by Pierre Teilhard de Chardin --- </br></br> .pull-left[ <div style="font-size:150%"> <i>[A] curious aspect of the theory of evolution is that everybody thinks he understands it.</i> <div style="text-align:right;"> — Jacques Monod</div> </div> ] .pull-right[  ] ??? Jacques Monod, Nobel prize for physiology or medicine, 1965 for his work on enzymes; what we would call today regulation of gene expression --- ## What is evolution? ✪ .pull-left[ We *observe* evolution: * change of species composition in geological time * unity of life – tree of life * shared genetic code * shared L-amino acids, protein structures, even complex enzymes * sequence conservation (even in neutral positions) * experimental evolution Theory of evolution is any theory that explains the observed evolution. ] .pull-right[  Ernst Haeckel, *Amaltheus margaritatus* (Ammonite) ] --- ## Short history of theory of evolution .pull-left[ Milestones: * in the XVIII century it became clear that the age of Earth is at least hundreds of thousands years, if not hundreds of millions (todays estimate: four billions) * XVIII/XIX century: first "trees of life" (Buffon) * XIX century: several theories (including Lamarck) – mechanism lacking! * crucial idea of natural selection: William Wallace, William Charles Welles, Patrick Matthew ] .pull-right[  ] --- ## Oh yeah, and Charles Darwin .pull-left[  ] .pull-right[  ] --- ## The principle of natural selection ✪ Natural selection is a *mechanism*: * IF an entity replicates: 1. with errors 2. these errors influence replication 3. and are heritable (heritability, variability, selection) -- * THEN: * change of phenotype over time will be observed --- ## The theory of natural selection ✪ .pull-left[ **Short summary of the theory of natural selection**: Natural selection is one of the main mechanisms responsible for evolution and speciation (hence, "Origin of the Species") ] .pull-right[  ] --- ## The problem with Darwin's theory .pull-left[ Darwin's theory was not complete: * Darwin did not know the mechanism of heredity * Darwin did not know about mutations ] -- .pull-right[ When studying fruit flies, Thomas Morgan (Nobel prize 1933) discovered mutations (e.g. white eyes) that were heritable. He showed that genes are located on chromosomes and that chromosomes are the mechanism of heredity. Earlier, he rediscovered mendelian genetics (Mendel's work was forgotten for decades). Thus, genetics was born. ] --- ## The great synthesis / modern synthesis ✪ .center[ <div style="font-size:200%"> Natural selection + Mendelian genetics + Neutral evolution = Modern synthesis </div> ] --- ## Remember! ✪ .center[ Evolution `\(\neq\)` Theory of Evolution Natural selection `\(\neq\)` Theory of Natural Selection Theory of Natural Selection `\(\neq\)` "The Great Synthesis" ] --- ## Other mechanisms of evolution ✪ * Genetic drift / neutral evolution * many mutations, possibly the majority (as we have seen in the multiple sequence alignments) have little or no impact on the structure / function * random propagation in small populations may "fix" alleles not through selection, but just by chance -- * Gene flow – exchange of alleles between populations --- ## Lactose intolerance / lactase persistence  ---  .myfootnote[ *Tishkoff et al. “Convergent adaptation of human lactase persistence in Africa and Europe.” Nat Genet. 2007 January; 39(1): 31–40; http://www.ncbi.nlm.nih.gov/pubmed/17159977.* ] --- .center[  **I am a mutant with superpowers.** ] -- .center[ *I don't fart after drinking milk *and* I don't suffer from vitamin D deficiency in high geographic latitudes.* ] --- ## Positive / negative selection ✪ -- .pull-left[ **Negative (purifying) selection** * non-neutral mutations are disadvantageous * results in highly conserved sequences * e.g.: * replication / transcription / translation * histones ] -- .pull-right[ **Positive (adaptive) selection** * non-neutral mutations are advantageous * therefore the sequences change rapidly * e.g.: * immune related genes (MHC) * Darwin's finches * Lactase persistence ] --- ## Genetic drift ✪ ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  --- ## Sequence evolution ✪ * synonymous vs non-synonymous mutations --- ## dN / dS (Ka/Ks) .pull-left[ Nucleotide: AAG ACT GCC GGG CGT ATT AAA ACA GCA GGA CGA ATC * * * * * * = 6 mutations Amino acid: K T A G R I K T A G R I = 6 synonymous, 0 non-synonymous ] .pull-right[ Nucleotide: AAG ACT GCC GGG CGT ATT AAA ACA GAC GGA CAT ATG * * * * * * = 6 mutations Amino acid: K T A G R I K T D G H M * * * = 3 synonymous, 3 non-synonymous ] --- ## Ka and Ks **Non-synonymous substitution rate:** `$$K_a = \frac{N_d}{N}$$` where `\(N_d\)` – number of non-synonymous substitutions, and `\(N\)` – number of nonsynonymous "sites" **Synonymous substitution rate:** `$$K_s = \frac{S_d}{S}$$` where `\(S_d\)` – number of synonymous substitutions, and `\(S\)` – number of synonymous "sites" Then **selective strength** `$$\omega = \frac{K_a}{K_s}$$` The devil is in the detail: how to calculate the number of synonymous sites? --- Consider the third site of the codons `AAA` and `AAG`. There exist a number of possible mutations: ``` AAA <-> AAC K <-> N NS AAA <-> AAG K <-> K S AAA <-> AAT K <-> N NS AAC <-> AAG N <-> K NS AAC <-> AAT N <-> N S AAG <-> AAT K <-> N NS ``` which makes 4 potential NS and 2 S mutations at this site. Also, we need to consider different models of evolution. The calculations quickly become complex. --- ## Ka/Ks ratio ✪ .pull-left[ **High Ka/Ks ratio:** * More non-synonymous mutations than expected: proteins evolve faster than by molecular clock alone. * Selection is positive / directional, pushing changes * Typical for genes that „are being selected“ (HLA/MHC, receptors) ] .pull-right[ **Low Ka/Ks ratio:** * Fewer non-synonymous mutations than expected: proteins evolve slower than by random mutations. * Selection is negative / stabilizing, maintaining current sequence * Typical for most functional sites, especially these which are functionally important (replication / transcription / translation, core metabolism) ] --- ## Diffent mutation rates in different proteins  --- class:empty-slide,myinverse background-image:url(images/portjacksonshark.jpg) ??? Port jackson shark --- class:empty-slide,myinverse background-image:url(images/heterodontus_zitteli.jpg) ??? Basically the same shark, but from the Jurassic (Berlin Museum of Natural History) --- ## Constant rate of mutation in hemoglobin * Heterodontiformes known from the Jurassic * number of differences between hemoglobin `\(\alpha\)` and hemoglobin `\(\beta\)` same as in humans, despite almost no changes in the phenotype --- ## Amino acid replacement rate almost constant  --- ## Neutral evolution ✪ * most of the mutations are neutral (even if non-synonymous!) * overall, the mutation rate can be quite constant over large periods of time `\(\rightarrow\)` no selection, "molecular clock" ---  .myfootnote[ *Ohta, “Near-neutrality in evolution of genes and gene regulation”, 2002. PNAS 99(25):16134-7* ] --- 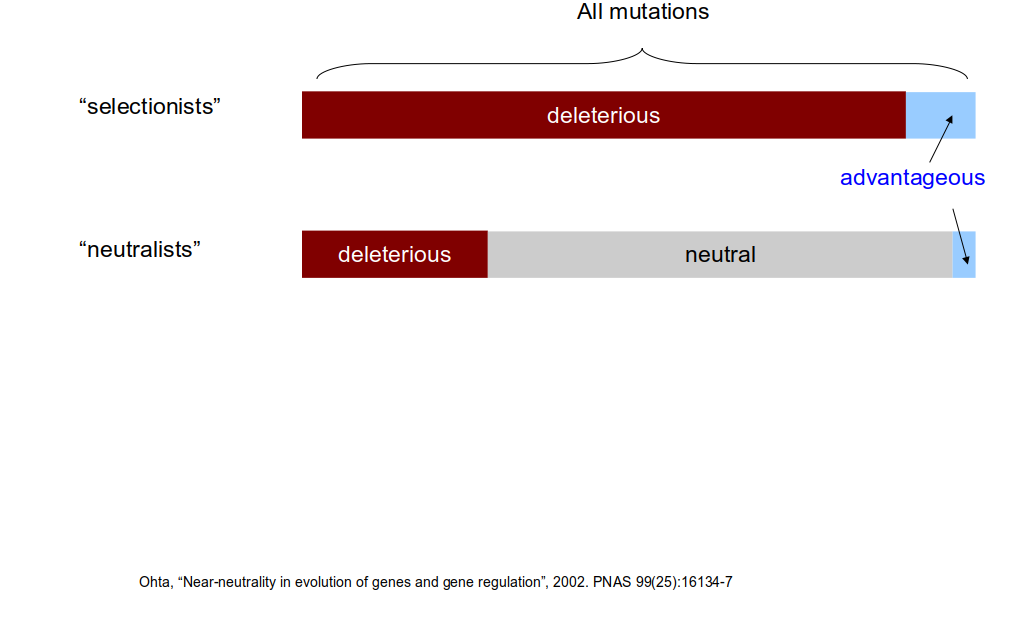 .myfootnote[ *Ohta, “Near-neutrality in evolution of genes and gene regulation”, 2002. PNAS 99(25):16134-7* ] --- 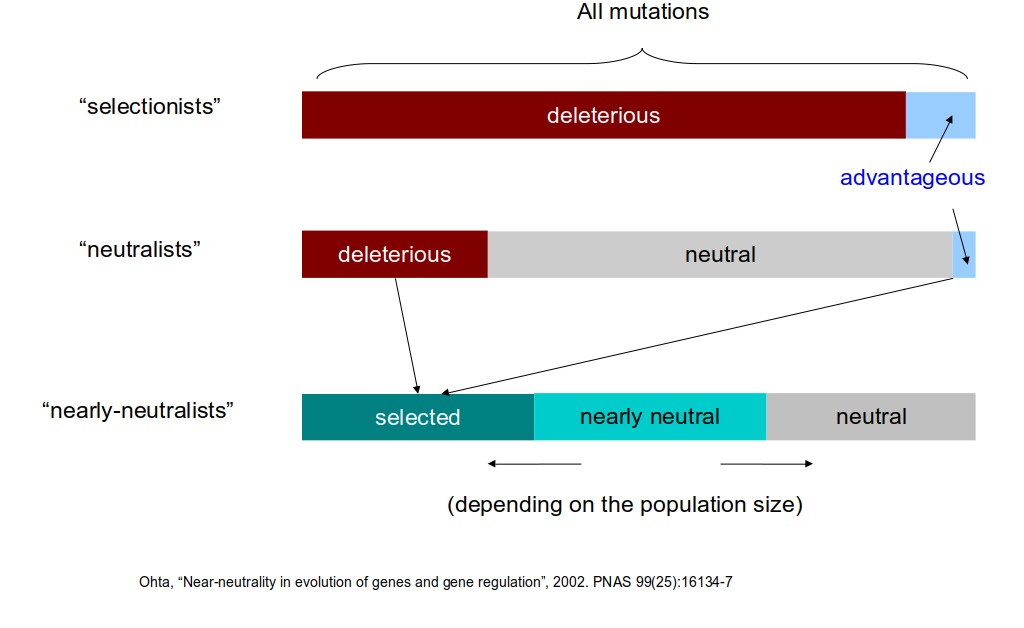 .myfootnote[ *Ohta, “Near-neutrality in evolution of genes and gene regulation”, 2002. PNAS 99(25):16134-7* ] ??? How to concile neutral evolution and selection? --- ## Was Darwin a selectionist¹? *I am inclined to suspect that we see, at least in some [cases], variations which are of no service to the species, and which consequently have not been seized on and rendered definite by natural selection.… **Variations neither useful nor injurious would not be affected by natural selection, and would be left either a fluctuating element**, as perhaps we see in certain polymorphic species, or would **ultimately become fixed**.… We may easily **err in attributing importance to characters, and in believing that they have been developed through natural selection**;… many structures are now of no direct use to their possessors, and may never have been of any use to their progenitors.…* <span style="text-align:right;">– Charles Darwin, The Origin of Species</span> .myfootnote[ ¹ Betteridge's law of headlines: "Any headline that ends in a question mark can be answered by the word no." ] --- ## A crash-course in phylogenies Why do phylogenetic reconstructions? * Answer exciting scientific questions * Create beautiful plots --- ## What makes a phylogenetic tree ✪ 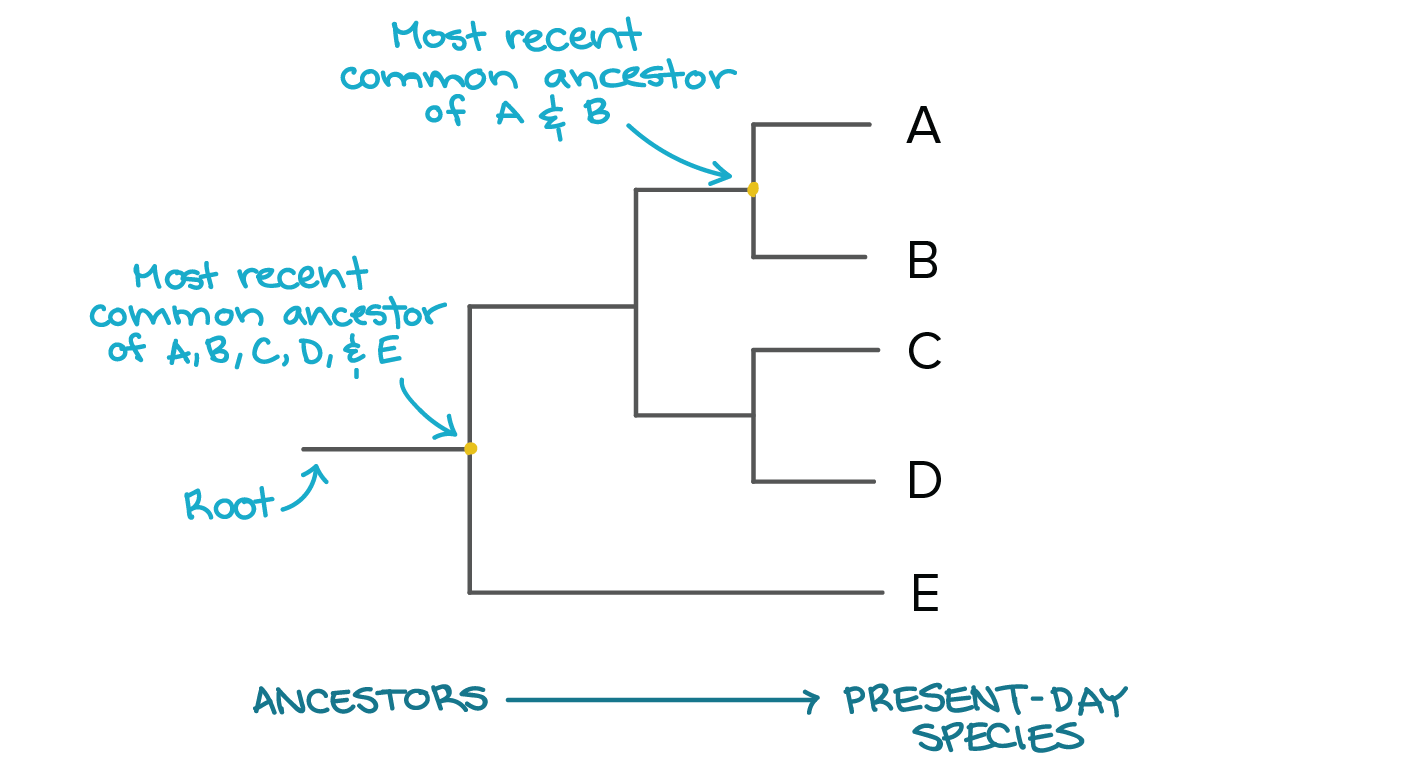 --- ## Rooted vs unrooted trees ✪ .pull-left[ 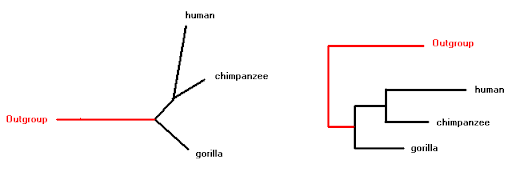 ] .pull-right[ To *root* a tree, we need an *outgroup* – a species for which we know for sure is not part of the group we are studying. For example, if we are studying primates, we can use a mouse as an outgroup. ] --- ## Tree Of Life .pull-left[  ] .pull-right[ *Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y. A new view of the tree of life. Nature microbiology. 2016 Apr 11;1(5):1-6.* ] --- ## Tree Of Life .center[  ] .myfootnote[ Interactive Tree of Life, [https://itol.embl.de/](https://itol.embl.de/) ] --- ## Tree Of Life .center[  ] .myfootnote[ Interactive Tree of Life, [https://itol.embl.de/](https://itol.embl.de/) ] --- ## Three domains of life .center[  ] --- ## A crash-course in phylogenies Why do phylogenetic reconstructions? * Answer exciting scientific questions * Create beautiful plots Also, practical applications: * Diseases * Functional annotation of genes by orthology reconstruction * Criminal prosecution -- ...wait, criminal prosecution? --- .pull-left[  ] .pull-right[  ] .myfootnote[ *Metzker, Michael L., et al. "Molecular evidence of HIV-1 transmission in a criminal case." Proceedings of the National Academy of Sciences 99.22 (2002): 14292-14297.* ] --- ## Drawing trees .pull-left[ * rooted vs unrooted * outgroup: used to root a tree * types of rooted trees ] --- ## Tree reconstruction  --- ## Tree reconstruction ✪ * Distance based methods: * calculate evolutionary distances (e.g. number of substitutions) between each two sequences * use the distances to reconstruct a tree * Parsimony based methods * Find the tree with the lowest number of evolutionary events (mutations, indels) * Maximum likelihood methods * find the tree with the highest likelihood: most likely to explain the observations * Bayesian methods * find the tree with the highest posterior probability --- ## Example: UPGMA ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  --- # Neighbor joining .pull-left[ * very similar to UPGMA * in each step: * based on the scores, calculate the "Q matrix" which can be understood as distance between two species *in relation* to their distances to other species * join the two species with the smallest Q value * recalculate the distances ] .pull-right[  ] --- Calculation of the Q matrix: `$$q_{i,j} \stackrel{def}{=} d_{i,j}\cdot (n-2) - \sum_{k \ne i, k \ne j} d(i, k) - \sum_{k \ne i, k \ne j} d(j, k)$$` Where `\(d_{i,j}\)` is the distance between nodes `\(i\)` and `\(j\)`. Explanation: basically, we look how far away are `\(i\)` and `\(j\)` *in relation* to other nodes. For example, for some node `\(k\)`, we can calculate `\(d_{i,j} - d_{i,k} - d_{j,k}\)`. The smaller this term will be, the closer `\(i\)` and `\(j\)` are to each other *in relation* to `\(k\)`. The `\(n - 2\)` is the number of remaining nodes. --- Recalculating the distances: When a new node is created (say, node `\(u\)`) from nodes `\(i\)` and `\(j\)`, we need to calculate first the distances of `\(i\)` and `\(j\)` to this new node: `$$d_{i,u} \stackrel{def}{=} \frac{d_{i,j}}{2} + \frac{\sum_{k=1}^n d_{i, k} - \sum_{k=1}^n d_{j, k}}{2\cdot n \cdot (n-2) }$$` We also need the new distances from that node to the remaining nodes: `$$d_{u, k} \stackrel{def}{=} \frac{(d_{i, k} - d_{i, u}) + (d_{j, k} - d_{j, u})}{2}$$` --- ## Final remarks This is just the beginning... * In this lecture, we do not go further into evolutionary modelling (maximum likelihood methods, bayesian methods etc), and also into how exactly distances are calculated. * Maximum likelihood methods do not use distances at all, but use sophisticated statistical models of sequence evolution. * Bayesian models are similar to maximum likelihood methods, but incorporate prior knowledge about the tree. * Distance based methods are very fast and they are still in use. * It is important to understand that the distances are calculated by assuming a certain model of evolution, including substitution rates. **When you create a phylogenetic tree, you always assume some model of evolution.**