class: center, middle, inverse, title-slide .title[ # Lecture 5: Reproducible Bioinformatics ] .subtitle[ ## BE_22 Bioinformatics SS 21 ] .author[ ### January Weiner ] .date[ ### 2024-05-06 ] --- ## Reproducibility crisis in science 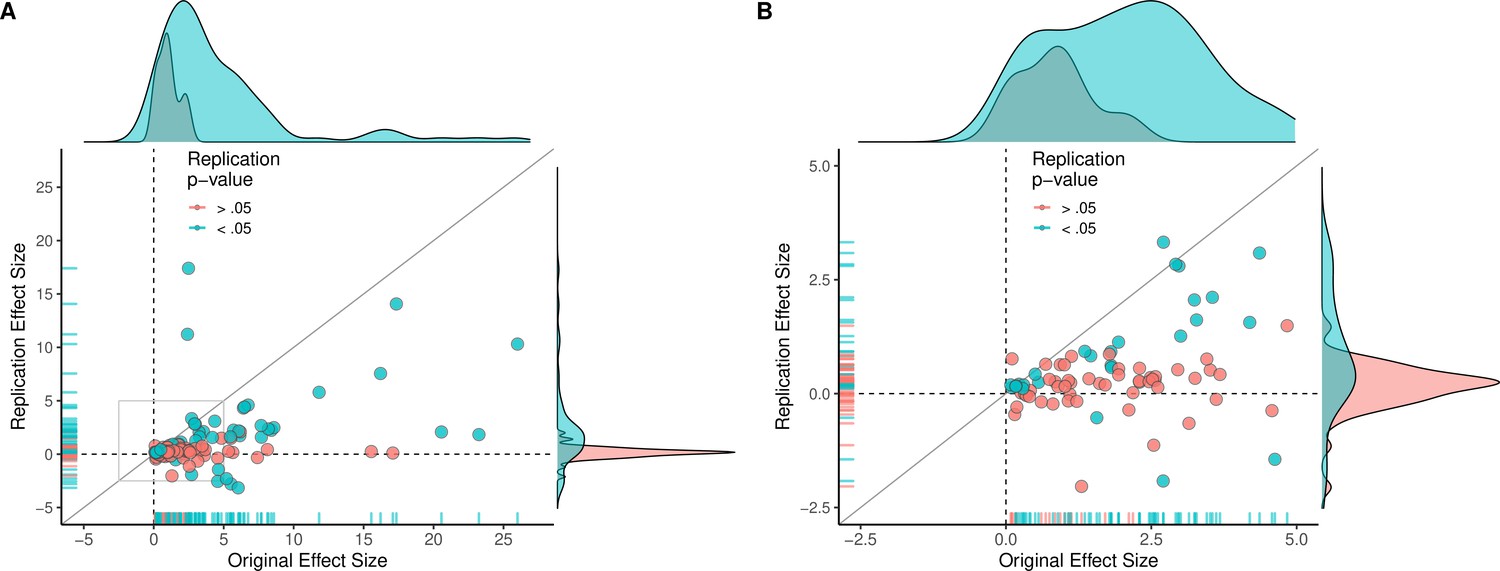 .myfootnote[ *Investigating the replicability of preclinical cancer biology TM Errington, M Mathur, CK Soderberg, …* - eLife, 2021 ] --- ## Reproducibility crisis in science 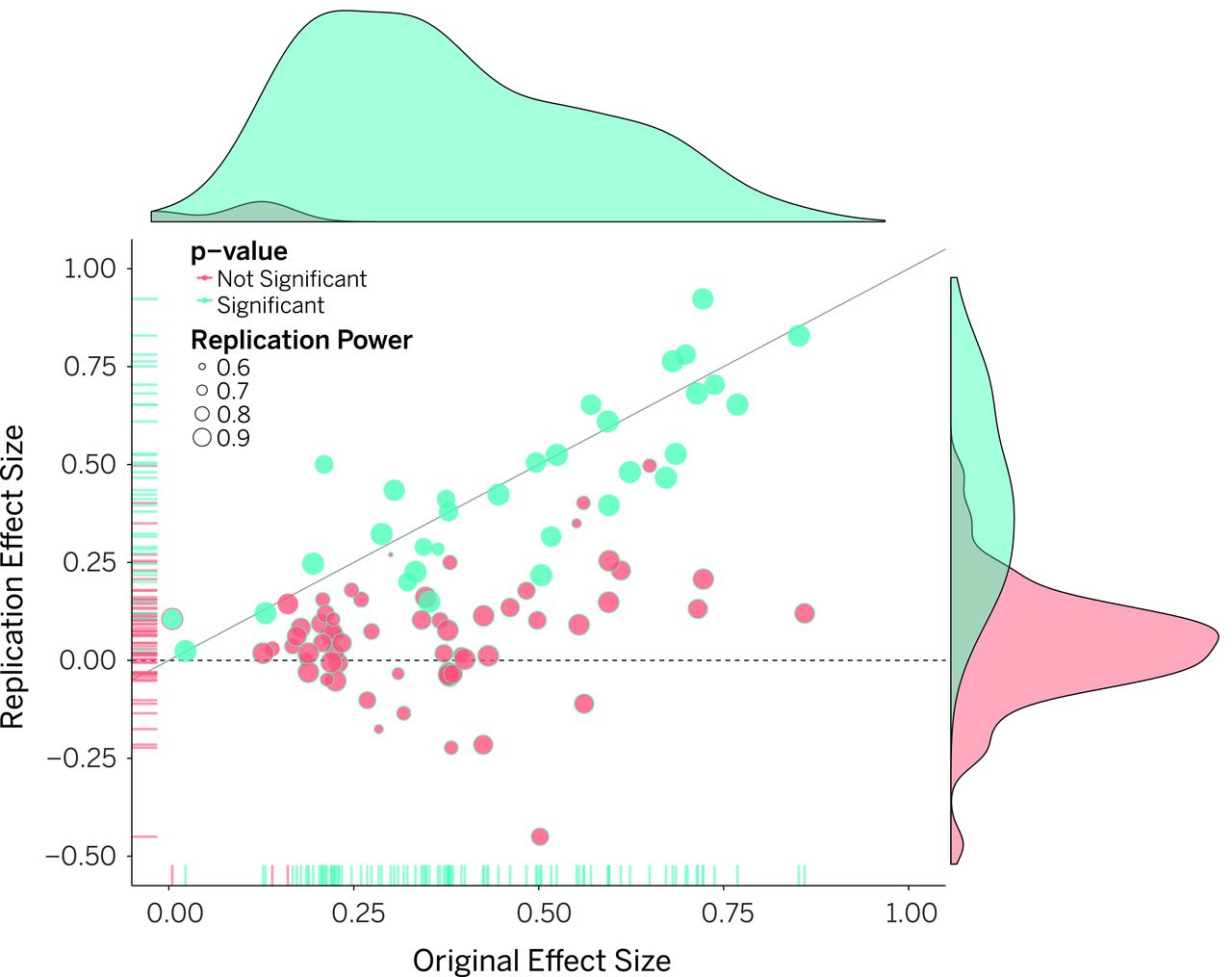 *Open Science Collaboration. "Estimating the reproducibility of psychological science." Science 349.6251 (2015).* --- ## Reproducibility crisis in science 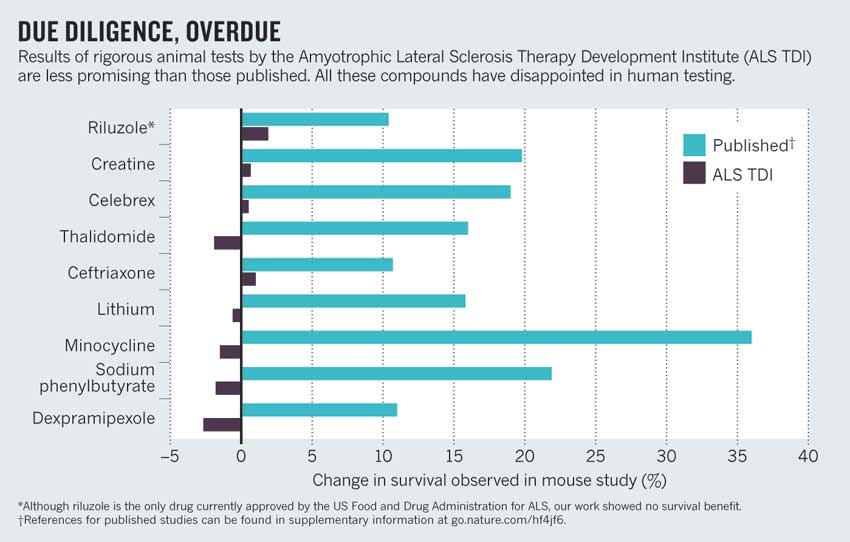 *Perrin, Steve. "Preclinical research: Make mouse studies work." Nature News 507.7493 (2014): 423.* --- ## Reproducibility crisis in science  --- ## Reproducibility crisis in science <table> <thead> <tr> <th style="text-align:left;"> Journal impact factor </th> <th style="text-align:right;"> Number of articles </th> <th style="text-align:left;"> Mean number of citations of non reproduced articles </th> <th style="text-align:left;"> Mean number of citations of reproduced articles </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> >20 </td> <td style="text-align:right;"> 21 </td> <td style="text-align:left;"> 248 (range 3–800) </td> <td style="text-align:left;"> 231 (range 82–519) </td> </tr> <tr> <td style="text-align:left;"> 5–19 </td> <td style="text-align:right;"> 32 </td> <td style="text-align:left;"> 169 (range 6–1,909) </td> <td style="text-align:left;"> 13 (range 3–24) </td> </tr> </tbody> </table> Bottom line: non-reproducible papers are more highly cited! --- 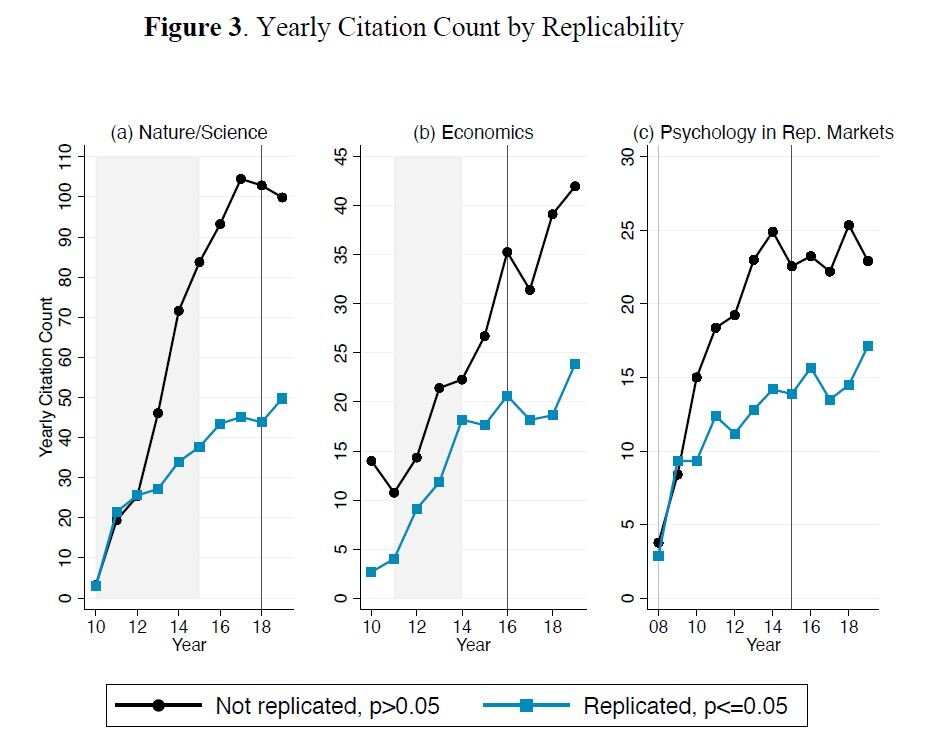 *source: https://phys.org/news/2021-05-replication-crisis-true-cited.html* --- ## Reasons for the reproducibility crisis: Money .center[  ] --- ## Reasons for the reproducibility crisis: Hype  --- ## Reasons for the reproducibility crisis: Lack of transparcency * no access to primary data * no precise description of methods: ``` We used generalized linear models (R package nlme4) to test the significance of association. ``` Fine, but how exactly was the model formulated? How many covariates? What transformations? What data were used? Which function? Were the results corrected for multiple testing? Etc., etc., etc. --- ## Reasons for the reproducibility crisis: Lack of transparcency .pull-left[ "...I thought it necessary to deliver things circumstantially, that the person I addressed them to might, without mistake, and with as little trouble as possible, be able to repeat such unusual experiments" *Robert Boyle, 1627-1691* ] .pull-right[  ] --- .myfootnote[ *Reproducibility in Cancer Biology: Challenges for assessing replicability in preclinical cancer biology* TM Errington, A Denis, N Perfito, E Iorns, BA Nosek - eLife, 2021 ] "The initial aim of the project was to repeat 193 experiments from 53 high-impact papers (...) [W]e were only able to repeat 50 experiments from 23 papers." -- "Second, <span style="background-color:yellow;">none of the 193 experiments were described in sufficient detail in the original paper to enable us to design protocols to repeat the experiments</span>, so we had to seek clarifications from the original authors." --- ## Of mice and men  --- ## Of mice and men .pull-left[  ] .pull-right[ * No or little correlation between transcriptomic responses in mouse models and human models: `\(r^2 < 0.1\)` – less than 10% of variance explained * No similarity in identified functions ] --- ## Of mice and men  --  --- ## Of mice and men .pull-left[  ] .pull-right[ * Strong correlation between mouse and human transcriptomic responses for genes which are regulated in one or the other condition: `\(\rho \geq 0.5\)`; change direction correlated * Similar functions for similarly regulated genes ] ??? None of the two papers described their methods sufficiently. --- ## By the way: about blinding...  --- ## Reasons for the reproducibility crisis: Statistics .center[  ] --- ## Hypothesis testing in science A primer on scientific method: -- *Kill your darlings* -- Rather than trying to *prove* your hypothesis, try to *disprove it*. Only a hypothesis which resists repeated attempts to overthrow it is useful. --- ## Statistical hypothesis testing A primer on statistical hypothesis testing: You create two models which can explain your data. One you call `\(H_0\)`, the other one `\(H_1\)`. -- The models differ by one tiny assumption, e.g. that the groups considered differ. -- Now you ask the question: which of the models is better? --- ## Statistical hypothesis testing Most frequently, we test this as follows: * we take the first model, `\(H_0\)`, and calculate the probability of generating our data using that model -- * if this probability is very low, we conclude that the model is not sufficient to explain the observed data -- * therefore we reject it in favor of the other model. --- ## Statitical hypothesis testing in science ✪ * Both `\(H_0\)` and `\(H_1\)` make certain assumptions like normality. The only difference is in the effect; usually, `\(H_0\)` assumes no effect, `\(H_1\)` assumes that there is an (unknown) effect * The reason why we use p-values calculated based on `\(H_0\)` is *that they are easy to calculate analytically* (i.e., without a fast computer). --- class:empty-slide,myinverse background-image:url(images/pregnant.jpg) --- ## What are p-values ...imagine we toss a coin... --- ## What are p-values Meaning of probability is different in classical (frequentist) statistics and in bayesian statistics * in frequentist statistics, this is the probability of getting a similar or better result assuming the `\(H_0\)`. `\(\Rightarrow\)` a coin that has landed is an event that happened, therefore p=1 OR p=0, but not p=0.5 * in bayesian statistics, it is a measure of information (much as our intuition tells us!) `\(\Rightarrow\)` as long as we don't know how the coin landed, we can use p=0.5 --- ## What are p-values p-values are defined within frequentist hypothesis testing. P-value gives us the average FPR (false positive rate, type I error rate) if we do many experiments, assuming the `\(H_0\)` ("no effect"). --- ## Reasons for the reproducibility crisis:<br>Misunderstanding and misusing of statistics ✪ -- * file drawer (suppresing non-significant results) -- * HARKing: Hypothesis After Results are Known -- * p-hacking / fishing / "researchers degrees of freedom" -- * "Garden of forking paths" (Gelman) --- ## Cherry-picking and p-hacking --- class:empty-slide,myinverse background-image:url(images/torturedata.jpg) ??? Stanisław Lem: "From strawberries under torture one may extract all sorts of things." --- class:empty-slide,myinverse background-image:url(images/vonneumann.jpg) --- ## Music makes people literally younger "*[W]e asked 20 University of Pennsylvania undergraduates to listen to either “When I’m Sixty-Four” by The Beatles or “Kalimba.” Then, in an ostensibly unrelated task, they indicated their birth date (mm/dd/yyyy) and their father’s age. We used father’s age to control for variation in baseline age across participants*. *An ANCOVA revealed the predicted effect: According to their birth dates, **people were nearly a year-and-a-half younger after listening to “When I’m Sixty-Four” (adjusted M = 20.1 years) rather than to “Kalimba” (adjusted M = 21.5 years), F(1, 17) = 4.92, p = .040**.*" .myfootnote[ *Simmons JP, Nelson LD, Simonsohn U. False-positive psychology: Undisclosed flexibility in data collection and analysis allows presenting anything as significant. Psychological science. 2011 Nov;22(11):1359-66.* ] --- ## A short guide to p-hacking * p > 0.05? Try another test * Or maybe add or remove some covariates? * Surely, there are outliers. Remove them. * Well, let us collect some more data. And then some more. As long as it takes. --- ## Evidence for p-hacking: the p-curve .pull-left[  ] .myfootnote[ left: *Head ML, Holman L, Lanfear R, Kahn AT, Jennions MD. The extent and consequences of p-hacking in science. PLoS Biol. 2015 Mar 13;13(3):e1002106.* right: *Masicampo EJ, Lalande DR. A peculiar prevalence of p values just below 0.05. Quarterly journal of experimental psychology. 2012 Nov;65(11):2271-9.* ] -- .pull-right[  ] --- ## Reasons for the reproducibility crisis:<br>Keeping data private .pull-left[  ] .pull-right[ "*We requested just the images of western blotting membranes and I wonder how such images can contain novel information that is worth being kept confidential.*" ] .myfootnote[ *Miyakawa, Tsuyoshi. "No raw data, no science: another possible source of the reproducibility crisis." (2020). Molecular Brain 13:24.* ] --- ## p = 0.05 * Arbitrary threshold introduced (mostly) by R.A. Fisher * In perfect circumstances, guarantees a false positive rate of 1 in 20 ``` Q: Why do so many colleges and grad schools teach p = 0.05? A: Because that's still what the scientific community and journal editors use. Q: Why do so many people still use p = 0.05? A: Because that's what they were taught in college or grad school. ``` *Wasserstein, Ronald L., and Nicole A. Lazar. "The ASA's statement on p-values: context, process, and purpose." The American Statistician (2016).* --- ## Is p = 0.05 enough? Consider a test for a disease (like a test for Sars-Cov-2 infection). * Sensitivity 80%: that is, no more than 20% False Negatives (FNs) * Specificity 95%: no more than 5% False Positives (FPs) Imagine you are tested and that the result is positive. What is the probability that you are infected with Sars-Cov-2? (**Note:** in reality, the Sars-Cov-2 tests are *much* more sensitive and specific) --- ## Is p = 0.05 enough?  --- ## Is p = 0.05 enough?  --- ## Is p = 0.05 enough?  --- ## Is p = 0.05 enough? OK, now replace "Sars-Cov-2 test" with "hypothesis test". * 80% sensitivity is normal for statistical tests (80% power) * 95% specificity corresponds to our p = 0.05 How many (scientific) hypotheses we are testing are really correct? * Surely much less than 100%, if it was close to 100% we would not be using statistics! * But more than 0%, we would not be doing science otherwise * Say, it is 10% (maybe 30% on a good day, but maybe 1% on a bad one). --- ## Is p = 0.05 enough?  --- .center[ <!-- --> ] --- .center[ <!-- --> ] --- class:empty-slide,myinverse background-image:url(images/dogs.jpg) --- ## Effect sizes: Does astrology work? Astrology: date of birth can be used to predict our physical and mental abilities. Can date of birth be used to predict our height? .myfootnote[ Weber, Gerhard W., Hermann Prossinger, and Horst Seidler. "Height depends on month of birth." Nature 391.6669 (1998): 754-755. ] ??? paper: https://www.nature.com/articles/35781 Weber, Gerhard W., Hermann Prossinger, and Horst Seidler. "Height depends on month of birth." Nature 391.6669 (1998): 754-755. --- ## Effect sizes: Does astrology work? .center[  ] --- ## Effect sizes: Does astrology work? .center[  ] --- ## Effect sizes: Does astrology work? * The observed effect was in the range of 6 mm * The standard deviation for human population is ~ 70 mm * Other factors (genetic, nutrition) have a much greater effect * Simulated comparison (N=100000): .center[ <!-- --> ] --- ## Effect sizes: Does astrology work? * The observed effect was in the range of 6 mm * The standard deviation for human population is ~ 70 mm * Other factors (genetic, nutrition) have a much greater effect * Simulated comparison (N=100000), as a boxplot: .pull-left[ <!-- --> ] .pull-right[ T-test results: <pre class="r-output"><code>## ## Welch Two Sample t-test ## ## data: df$Height by df[[2]] ## t = 19.061, df = 2e+05, p-value < 2.2e-16 ## alternative hypothesis: true difference in means between group April and group October is not equal to 0 ## 95 percent confidence interval: ## 0.5343158 0.6567946 ## sample estimates: ## mean in group April mean in group October ## 177.2962 176.7006 </code></pre> `\(\Rightarrow\)` Differences are statistically significant! ] --- ## Bottom line ✪ * p-value is a part of scientific *language*. Participating in scientific communication (reading and writing papers) is dangerous if you do not understand what it means. -- * p-values never tell us about the probability of `\(H_1\)` -- * p-values are meaningless when we reject `\(H_0\)`! They refer to an alternate universe in which we assume that `\(H_0\)` is true -- * p-values can be "extremely significant" (incorrect phrase!), but the effect may still be very small. "Statistically significant" is not the same as "significant". -- * p-values are calculated given certain assumptions (the null hypothesis, `\(H_0\)`), and not "randomness". This can lead to errors if these assumptions are not met. Examples: independence, normality -- * Absence of evidence (i.e. significance) is not evidence of absence (of the effect). That means: lack of statistical significance is not *evidence* that there is no effect. -- * p-value is a *tool*. Like any tool, it can be incorrectly used. It will not make a decision for you. -- .center[ <div style="font-size:150%">**THIS. SHIT. IS. IMPORTANT.**</div> .] --- ## Correlation and causation ✪ .center[  ] --- ## Correlation and causation ✪ .center[  ] --- ## Correlation and causation ✪ .center[  ] --- ## Correlation and causation * The Japanese eat very little fat and suffer fewer heart attacks than the British or Americans. -- * The French eat a lot of fat and also suffer fewer heart attacks than the British or Americans. -- * The Japanese drink very little red wine and suffer fewer heart attacks than the British or Americans. -- * The Italians drink excessive amounts of red wine and also suffer fewer heart attacks than the British or Americans. -- * The Germans drink a lot of beer and eat lots of sausages and fats and suffer fewer heart attacks than the British or Americans. -- **Conclusion:** Eat and drink what you like. Speaking English is what kills you. -- (actually: most of the facts above are **untrue!**) --- ## Correlation and causation Say, you observe that people who express certain behavior (**B**) also tend to succumb to a certain disease (**D**). Several explanations are possible: * **B** causes **D** * There is a common factor that causes both **D** and **B** * Predisposition to **D** causes **B** * Predisposition to **B** is linked with predisposition to **D** Should we dismiss **B** as a cause for **D**? --- class:empty-slide,myinverse background-image:url(images/cigarettes.png) --- ## Correlation and causation .pull-left[  Ronald A. Fisher ] .pull-right[ "Is it possible, then, that lung cancer—that is to say, the pre-cancerous condition which must exist and is known to exist for years in those who are going to show overt lung cancer—is one of the causes of smoking cigarettes? I don't think it can be excluded." (Fisher RA. *Cigarettes, cancer and statistics*. Centen Rev 1958;2:151-66.) ] --- ## Multiple testing * Every time you run a statistical test, you have a chance of fooling yourself * In bioinformatics, running tens of thousands of statistical tests is pretty common --- class:empty-slide,myinverse background-image:url(images/salmon_1.png) --- ## Multiple testing .pull-left[ **Subject.** One mature Atlantic Salmon (Salmo salar) participated in the fMRI study. The salmon was approximately 18 inches long, weighed 3.8 lbs, and was not alive at the time of scanning. **Task.** The task administered to the salmon involved completing an open-ended mentalizing task. The salmon was shown a series of photographs depicting human individuals in social situations with a specified emotional valence. The salmon was asked to determine what emotion the individual in the photo must have been experiencing. ] .pull-right[  ] --- ## What can we do as scientists? ✪ * registered studies, SOPs etc.: plan and publish the analysis before performing it * be as transparent as possible * publish data * publish methods as code on github (or similar) * learn statistics * KILL YOUR DARLINGS --- ## What can we do as non-scientists? ✪ * Beware of the hype * Always check the sources * Learn how to read scientific papers * Confront the sources (there is always another opinion) * Learn statistics --- ## Introduction to the report: rules ✪ * *In any case*, you will do the report in Rmarkdown or Jupyter lab book. * There are three main options: * Pick a protein and use any available tools to analyse its structure, evolutionary history (phylogeny), function etc. * Differential gene expression analysis, combined with functional analysis * Writing your own Python or R program (some ideas will follow) that actually does something useful * The main criterion is that your program or your Jupyter notebook or your Rmarkdown actually works and can produce a document (HTML, Word, whatever) --- ## Report on structural analysis * Pick a protein. Whatever you pick is OK, as long as you don't pick a protein or gene we have already discussed in the practicals * Use several of the following tools, or any other tools you find useful: * BLAST * Pfam * clustalw * phylogeny tools * InterPro * CATH * SCOP * PDB * UniProt * STRING * Try to apply these tools to the sequence of your protein to find out more about it * Document all things you have done in a *replicable* manner * Use the "Discussion" part to describe your opinion on the results, speculations, and what else one could do --- --- ## Report on differential gene expression analysis * Run a basic differential gene expression analysis as shown in the course * Create a visualisation of the results (volcano plot) * Run a gene set enrichment analysis * Select one of the top results and describe its structure *or* phylogeny * Document all things you have done in a *replicable* manner * Use the "Discussion" part to describe your opinion on the results, speculations, and what els could one do --- ## Report on your own program * document the program in your Rmarkdown document / Jupyter lab book (what does it do? What did you do with it? what were the results? what are the limitations?) * Document all things you have done in a *replicable* manner * Use the Discussion part to show what else could one do with the program, or how it could be expanded * It is more important that the program works and that you have documented it well than that is has all the features one could dream of --- ## Writing the report ✪ * With these tasks complete, you write a report * Report must be written in Rmarkdown or as Jupyter lab book * You must create a bibliography and include the references * Like a scientific paper, the report **must** include the following parts: * Abstract: short description of your work * Introduction: explain what is the background of your work (e.g. why analysing proteins is important) * Methods: describe, *in detail*, the methods you will apply. Cite sources! * Results: describe, *with figures*, your findings. * Discussion: what is your opinion? what else should / could one do? What did you learn? * Bibliography --- ## Writing the report ✪ There are three types of statements: * Statements by others: you must cite them. * Statements derived from your findings: you must back them up with results * Statements which are speculative: you must label them clearly as such