class: center, middle, inverse, title-slide .title[ # Lecture 1: What is bioinformatics? ] .subtitle[ ## BE_22 Bioinformatics SS 24 ] .author[ ### January Weiner ] .date[ ### 2024-04-05 ] --- ## Berlin Institute of Health .pull-left[ * Institute for translational and precision medicine * Founded in 2013 by Max Delbrück Center for Molecular Medicine and Charité – a university hospital affiliated with both Humboldt University and Free University Berlin. * Since 2020 a part of the Charité ]  ??? Charité – over 15,000 employes --- ## A few words about myself Short bio: * PhD in molecular biology * 2003-2009 evolutionary bioinformatics (sequence analyses, phylogeny, teaching) * 2009-2019 Max Planck for Infection Biology – tuberculosis, Omics, statistics, clinical trials * Since 2019 at the Core Unit for Bioinformatics, BIH, Charité Areas of expertise: * biostatistics, R * functional analysis * evolutionary biology --- ## Organisatory matters * Course is in English * Lectures: some live on Mondays, some pre-recorded and available on Friday before the Lecture * Practicals: * Fridays, 16-18 * 2 hours: guided exercise ("practice"): 1h45m with a 15' break * always a possibility to stay on longer and talk to me ??? what is the difference between this course and an online course from say Coursera? --- ## Lecture files * Lectures are in HTML format * Generated from Rmarkdown * Lectures are on github [https://github.com/january3/Bioinformatics](https://github.com/january3/Bioinformatics) * See [keyboard shortcuts](https://bookdown.org/yihui/rmarkdown/xaringan-key.html) * Important slides / concepts marked with ✪ --- ## Practicals * First part: using online tools for fun and profit * Second part: learn how to program using R --- ## Course evaluation ✪ * Attendance is *not* mandatory * Homeworks (short and easy) 30% = 6 x 5% * Report ("paper") 30% * Exam (multiple choice) 40% * Additional 15 points for active participation in the course Rule #1: no plagiarism --- ## Homeworks * easy and quick to do * you will have at least 4 weeks for each homework * must be done within the deadline (to reward those who work systematically) * must be submitted via Moodle --- ## Reports * Must be written as an Rmarkdown document, plus output (html or docx or pdf), or a Jupiter notebook * Must be your own work * The actual work will be simple * I will not be evaluating correctness of the solutions or code, but * whether you did all that you were ask to do * document structure * bibliography * You are allowed any programming language I know. This includes R, Python, bash, C, C++, Java, Perl, Basic, matlab and lua. If you prefer another language, contact me. * Detailed instructions (will be) on Moodle --- ## What about AI (ChatGPT, Bing search, copilot...) .pull-left[ Use it! It's the new Wikipedia, Google, Stackoverflow, Github. But... * Know its limits * Understand how it works * Never. Ever. Fully trust it. It LIES. ] --- .pull-left[  ] .pull-right[  ] --- ## Rules on using ChatGPT and other LLMs LLMs *are* useful. I use them all the time. However, as any powerful tool they can be dangerous! * You may use LLMs for: * finding information * explaining concepts * learning how to program * finding errors in your code * finding errors in your language * You are **absolutely not allowed to** use LLMs to write your reports, homeworks or exams for you. This is plagiarism and will be punished as such. * Keep in mind that although detecting LLM output is harder than detecting regular plagiarism, there *are* tools that can do that. --- ## How to work safely with LLMs such as ChatGPT .pull-left[ My advice: * **NEVER, EVER** include any sensitive or copyrighted data in your queries. Basically, assume that anything you write will be published on the Internet. * Always check the output for errors * Always check the output for plagiarism * Generally, popular topics / tools are safer to learn from LLMs; obscure or advanced methods are more likely to be (completely) wrong * Never copy-paste the output directly into your report or your code * **NEVER, EVER** use the output which you don't understand * better: discuss with the LLM the concepts you don't understand * write your own code based on the LLM output * check with LLM in case of problems ] .pull-right[  ] LLMs can be your personal trainer - or the reason for your failure. --- .pull-left[  ] .pull-right[  ] --- ## Why are you learning bioinformatics? * Because you must? -> then at least don't waste your time, chances are you will learn something useful -- * Because you cannot work without computers? -> you will have to use them -- * Because it is interesting? -> welcome! -- There is no correct answer. .pull-left[ Personally, I like bioinformatics, because: * It allows me to work with many different scientific topics and many different scientists - it is never boring * Coding (programming) is fun, and science is fun, and bioinformatics is both * I like statistics, and bioinformatics can be a lot of statistics ] -- .pull-right[ I also hate bioinformatics, because * Coding can be frustrating * Software engineering can be really boring (maintaining code, debugging, user support) * It is hard to have your own scientific topic without being able to run your own experiments * Keeping up with new developments takes up a lot of time ] --- ## General advice 1. Ask questions. -- 2. Do **ASK** questions. -- 3. Take notes, don't rely on lecture materials. -- 4. Try things on your own. --- ## Let me tell you a story --- class:empty-slide,myinverse background-image:url(images/Slum-pic-1-Wikipedia.jpg) ??? London in the 1850s: Irland immigration, slums, exponential population growth, from 1 million in 1800 to over 2 in 1850 and 4 in 1900 --- class:empty-slide,myinverse background-image:url(images/Slum-pic-4.jpg) ??? The rookeries - the worst slums in London --- class:empty-slide,myinverse background-image:url(images/Slum-pic-3.jpg) ??? "2-penny hangover" - fake picture, though --- ## London cholera epidemics * 1831-32: 30 thousand deaths * 1850-52: 50 thousand deaths * 1853: 10 thousand deaths --- ## The miasma theory of cholera  ??? Florence Nightingale, William Farr (regarded as the father of medical statistics) --- ## Alternative explanation .pull-left[ **John Snow** – 1813-1858, physycian, early anaesthesiologist ] -- .pull-right[  ] --- ## Alternative explanation .pull-left[ **John Snow** – 1813-1858, physycian, early anaesthesiologist Observations: * cholera is linked to the digestive tract * it appears that it is contagious Speculations: * the "cholera poison" must somehow multiply * possibly it multiplies in the digestive tract * faeces contaminates drinking water with "cholera poison" However: * In 1831, two German physicians tried to test contagiousness by drinking faeces of a cholera patient * epidemies come and go, despite of the water still being contaminated ] .pull-right[  ] --- ## Testing the hypothesis * Collecting cases in which neighborhoods differed with respect to cholera outbreaks, predicting that they would have a different water supply * This required testing the water to determine the supplier * Also suggested switching the supplier, which had an effect on the epidemic * Used William Farrs data on cholera epidemics ??? two water suppliers had particularly dirty water case when two "courts" one with epidemic second without, different water suppliers --- ## Broad Street epidemics In 1854, a serious outbreak occured in Soho (eventually, more than 600 people died all in the same neighborhood) --- ## Broad Street epidemics .pull-left[  ] .pull-right[ * Snow created a map combining geographical information with statistical data – deaths were represented as stack bars ] --- ## Broad Street epidemics .pull-left[  ] .pull-right[ * He then went on to include information on the nearest avaialble water source – a pump – by delineating boundaries between the regions close to one or another pump (this is called a Voronoi✪ diagram) ] --- ## Broad Street epidemics .pull-left[  ] .pull-right[ * He then went on to include information on the nearest avaialble water source – a pump – by delineating boundaries between the regions close to one or another pump (this is called a Voronoi✪ diagram) ] ??? Additionally, none of the workers who worked at a local brewery contracted cholera. Pump was then disabled by the authorities, but this is unlikely to be the cause of the end of the epidemics Later it was found that an old abandoned cesspit was leaking --- ## Broad street pump .pull-left[ * Board of Health attributed the outbreak to miasma * In 1854, italian scientist Filippo Pacini discovered the cholera bacterium, but *no-one noticed* * Snow died in 1858 due to a stroke * In 1866, William Farr realised during another outbreak that he was wrong after all (although he still did not believe in the "cholera poison") * In 1884, Robert Koch discovered and named *Vibrio cholerae*, showing that it causes cholera, and *everyone* noticed ] .pull-right[  ] ??? The problem most people had with the fecal-oral route --- ## The real cause of cholera .pull-left[  ] .pull-right[ Pacini discovered *Vibrio cholera* in 1854, 30 years before Koch did the same (and became famous for it).  ] --- ## Lessons from this story * That may have been a very early case of "bioinformatic analysis" * You may be right, but you need to convince others * You may be right, but still have no impact on science – you need to communicate it to the right crowd (other example: Gregor Johann Mendel) * An old theory is hard to kill * A good "story" helps (everybody knows about the pump) * A good visualization is not only for presenting data, but also for exploring it --- ## So, what is bioinformatics? .left[  ] .right[ *If bioinformatics was an animal...* ] --- ## Defining bioinformatics by the areas of research✪ (also, incidentally, plan for our course) -- .pull-left[ * Sequence analysis * Sequence alignments and sequence similarity * Sequence search * Motifs * Phylogenies and evolution ] -- .pull-right[ * Protein structures * Protein structure prediction * Visualizations ] -- .pull-left[ * Omics data analysis * Transcriptomics * Functional analysis ] -- .pull-right[ * Data science * Data management in biology * Data visualisation – a primer ] -- .pull-left[ * Machine learning * Basic concepts * Simple approaches ] --- ## Defining bioinformatics by tools of the trade✪ or, *How I became a bioinformatician* The easiest thing: programming skill * Python or R? * => Multilinguality pays off: learn at least the rudiments of HTML, bash, C, GNU make, snakemake to be able to install the tools of your trade * Learn how to efficiently use a terminal: use Linux / MacOS / Linux Subsystem for Windows (LSW) * Learn to use SSH, git, github / gitlab, markdown * Learn how to visualize data (with R or Python) * **Learn to touch-type, as you will be typing A LOT** --- ## Scientific skills * Bioinformatics is an area of science rather than technology * "Figure out how it works" rather than "build it"  --- ## Scientific skills * Bioinformatics is an area of science rather than technology * "Figure out how it works" rather than "build it"  --- ## Scientific skills✪ * Who is a scientist? * Read scientific papers * Learn how to write scientific papers * Understanding biology makes you a much better bioinformatician * Learn about scientific reasoning, hypothesis testing etc. .pull-right[  ] ??? Scientist is a person who reads *and writes* scientific papers. --- ## The art of fast reading of scientific papers * Search for papers using [google scholar](https://scholar.google.com) or [PubMed](https://pubmed.ncbi.nlm.nih.gov) * Use forward search! * Start with review articles, easier to read and will allow you to get into the subject * Most papers have Abstract, Introduction, Methods, Results and Discussion * Fastest method: look at the last sentence of an abstract, and maybe at the end of "Conclusions" if it is present * Second fastest: look at the figures * Third fastest: read the introduction and discussion, then check the results while they are discussed * If you want to *cite* a paper, you *must* read it in whole --- ## Important ('s why I use CAPSLOCK) * **DON'T TAKE THE AUTHORS BY THEIR WORD!** Try to find their errors / mistakes / omissions, ask "what if", or even "they must be joking" * **BE CAREFUL OF PRE-PRINTS, HERE BE DRAGONS** * **EVEN A NATURE PAPER MAY BE WRONG!**  --- ## Statistical skills * Statistics *is* hard. * Proper statistics is harder. * Science is in a reproducibility crisis, statistical methods are at the core of it. * Good bioinformatics means good reproducibility means good science .pull-right[  ] --- ## Story time 1. Biomarkers of tuberculosis (TB) progression 2. Genetic background in severe COVID-19 3. How Venn diagrams mislead scientists --- ## 1. Biomarkers of TB progression --- class:empty-slide,mywhite background-image:url(images/tuberculosis.png) ??? 1.4 mln deaths in 2019 10 mln new patients in 2019 400 thousand with MDR TB --- class:empty-slide,mywhite background-image:url(images/tuberculosis_2.png) --- class:empty-slide,mywhite background-image:url(images/study_scheme.png) --- Go to [manuscript](https://www.nature.com/articles/s41467-018-07635-7) --- ## 4. Transcriptomic responses in vaccination --- class:empty-slide,mywhite background-image:url(images/transcriptomics.png) --- ## ✪ How to make sens from thousands of results? .pull-left[  ] .pull-right[ * How do you make sense of a list of 1000 genes? * What if there are none, can you still say something? * What about missing functions? ] --- ## Gene set enrichments ✪ * Categorize genes into sets of related function * Instead of asking whether a single gene is regulated, ask whether the sets (as a whole) is regulated Advantages: * There are fewer sets than genes * Sets often have well understand biological functions ---  .footnote[*Zyla J, Marczyk M, Domaszewska T, Kaufmann SH, Polanska J, Weiner 3rd J. Gene set enrichment for reproducible science: comparison of CERNO and eight other algorithms. Bioinformatics. 2019 Dec 15;35(24):5146-54.*] ---  ---  .pull-left[Vaccine with an adjuvant] .pull-right[Vaccine without an adjuvant] .footnote[*Weiner, January, et al. "Characterization of potential biomarkers of reactogenicity of licensed antiviral vaccines: randomized controlled clinical trials conducted by the BIOVACSAFE consortium." Scientific reports 9.1 (2019): 1-14.*] --- ## How Venn diagrams can fool scientists  .footnote[*Venn diagrams may indicate erroneous statistical reasoning in transcriptomics. Weiner, Obermayer and Beule, 2022, Frontiers Genetics, in press*] --- ## What happens is, we are comparing significance with non-significance > The Difference Between “Significant” and “Not Significant” is not > Itself Statistically Significant - Andrew Gelman and Howard Stern (we will talk more about significance and lack of significance later!) --- Nieuwenhuis et al. found that half of the scientists who could have commited this error, did in fact commit this error.  .footnote[*Nieuwenhuis, Sander, Birte U. Forstmann, and Eric-Jan Wagenmakers. "Erroneous analyses of interactions in neuroscience: a problem of significance." Nature neuroscience 14.9 (2011): 1105-1107.*] --- ## The results are artifacts! Groups G1 and G2 were randomly drawn from the same population. They were not different at all!  --- ## 4. HLAs and COVID-19 severity ---  .footnote[*Once Upon a Time... Life*] --- ## Example of a real world study: HLAs and Covid .pull-left[ Is severe COVID-19 associated with a genetic background? 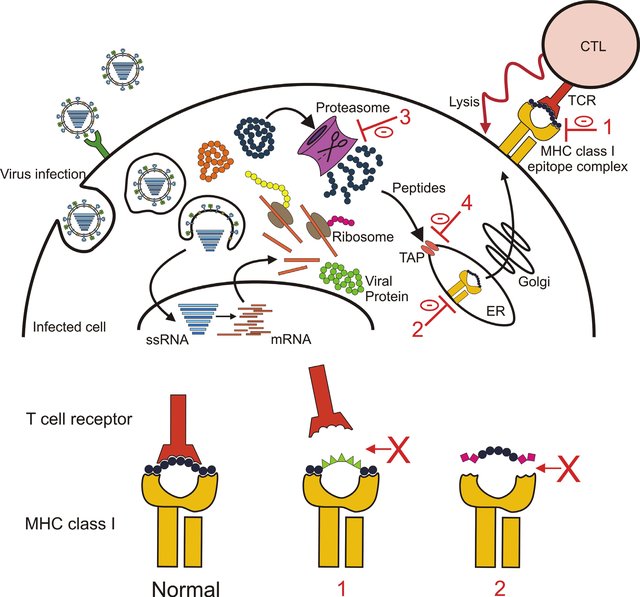 ] -- .pull-right[ HLA ("human leukocyte antigen") – molecule which binds and presents antigens to immune cells * There are different HLA genes * Each gene comes in different variants – alleles * Different alleles may be better or worse at "catching" an antigen * There is a huge variability of HLAs in humans (that is why it is so hard to transplant an organ) ] --- ## Genetic association testing ✪ * Test whether a *phenotype* (e.g. severe disease) occurs frequently in connection with a *genotype* (e.g. a particular allele) Sounds straightforward: * Take samples from COVID patients at the Charité * Determine their HLA alleles * Compare with who has been intubated and who was not .footnote[*Weiner 3rd, January, et al. "Increased risk of severe clinical course of COVID-19 in carriers of HLA-C 04: 01." EClinicalMedicine 40 (2021): 101099.*] --- ## Example of a real world study: HLAs and Covid 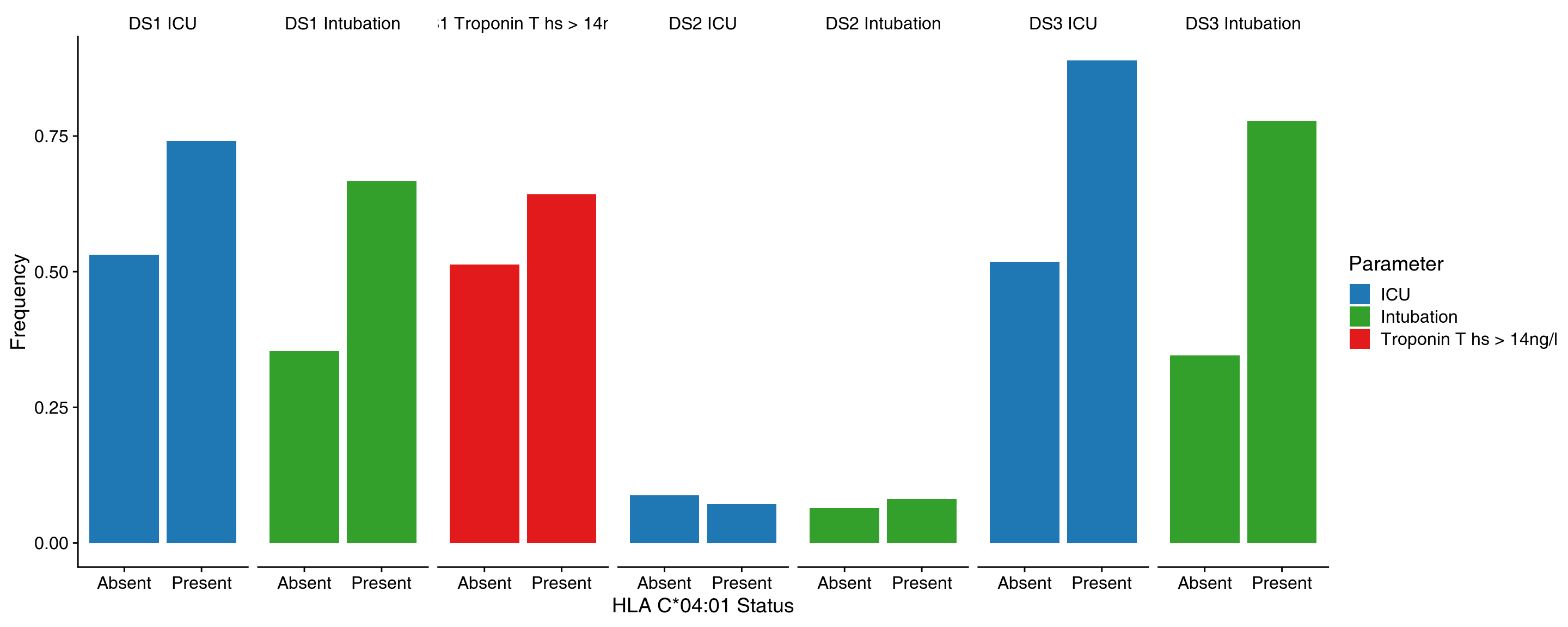 .footnote[*Weiner 3rd, January, et al. "Increased risk of severe clinical course of COVID-19 in carriers of HLA-C 04: 01." EClinicalMedicine 40 (2021): 101099.*] --- ## Example of a real world study: HLAs and Covid 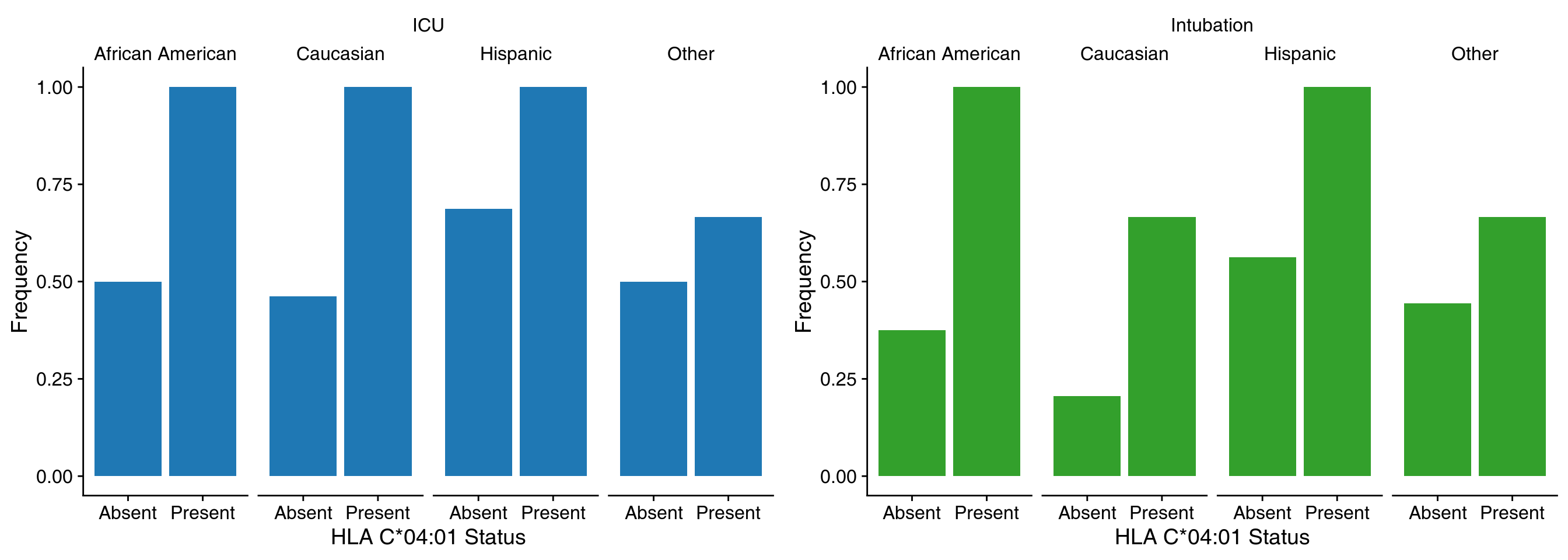 .footnote[*Weiner 3rd, January, et al. "Increased risk of severe clinical course of COVID-19 in carriers of HLA-C 04: 01." EClinicalMedicine 40 (2021): 101099.*] --- ## Real world vs Randomized Controlled trial (RCT) ✪ .pull-left[ "Real world" (observational) studies: * Compare existing groups: e.g. people who consume olive oil and people who do not. Does olive oil reduce cancer rates? * Problem: comparing apples to oranges * problems finding matching controls * unknown biases * unplanned comparisons * the cherry-picking Pandora box ] -- .pull-right[ RCT: * volunteers randomized in two groups * groups have now identical characteristcs * no bias due to group selection possible * planned comparisons prevent cherry-picking  *first RCT published – 1948* ] ??? - Fisher published his book in 1935 - Ethics: streptomycin was scarce anyway [@nieuwenhuis2011erroneous] --- # References <a name=bib-bmc1948streptomycin></a>[Streptomycin treatment of pulmonary tuberculosis](#cite-bmc1948streptomycin) (1948). "Streptomycin treatment of pulmonary tuberculosis". In: _British Medical Journal_ 2, pp. 769-782. <a name=bib-gelman2006difference></a>[Gelman, A. and H. Stern](#cite-gelman2006difference) (2006). "The difference between “significant” and “not significant” is not itself statistically significant". In: _The American Statistician_ 60.4, pp. 328-331. <a name=bib-nieuwenhuis2011erroneous></a>[Nieuwenhuis, S., B. U. Forstmann, and E. Wagenmakers](#cite-nieuwenhuis2011erroneous) (2011). "Erroneous analyses of interactions in neuroscience: a problem of significance". In: _Nature neuroscience_ 14.9, pp. 1105-1107. <a name=bib-weiner2021increased></a>[Weiner 3rd, J., P. Suwalski, M. Holtgrewe, et al.](#cite-weiner2021increased) (2021). "Increased risk of severe clinical course of COVID-19 in carriers of HLA-C* 04: 01". In: _EClinicalMedicine_ 40, p. 101099. <a name=bib-weiner2019characterization></a>[Weiner, J., D. J. Lewis, J. Maertzdorf, et al.](#cite-weiner2019characterization) (2019). "Characterization of potential biomarkers of reactogenicity of licensed antiviral vaccines: randomized controlled clinical trials conducted by the BIOVACSAFE consortium". In: _Scientific reports_ 9.1, pp. 1-14. <a name=bib-weiner2018metabolite></a>[Weiner, J., J. Maertzdorf, J. S. Sutherland, et al.](#cite-weiner2018metabolite) (2018). "Metabolite changes in blood predict the onset of tuberculosis". In: _Nature communications_ 9.1, pp. 1-12.